

- THE INTENSITIES OF BANDS WERE DETERMINED BY IMAGEJ SOFTWARE SOFTWARE
- THE INTENSITIES OF BANDS WERE DETERMINED BY IMAGEJ SOFTWARE SERIES
Run("Subtract Background.", "rolling=12") Masks will show the black and white image similar during thresholding.Įxclude Edges - ImageJ will not include cells that are not fully contained in the boundaries of the image Show - Outlines will trace the outside of the mask images. The minimum bound deviates from being a circle. The maximum bound (1.0) means a perfect circle. Size - parameter of what cells to include in data by area (pixels^2)Ĭircularity - parameter of what cells to include by how close to a circle the shape appears. Watershed - automated separation of separate fused cells by a 1 pixel lineĪnalyze Particles - process the image to acquire a cell count Threshold - change the image to binary image of red, black and white, or blueįill Holes - fill empty spaces between rings to make circlesĬonvert to Mask - allows for subsequent processing Subtract Background - cancel out noise of the background However, the error of missing cells may be "corrected" by the error of including some noise. Keep in mind that ImageJ processing is not perfect.
THE INTENSITIES OF BANDS WERE DETERMINED BY IMAGEJ SOFTWARE SOFTWARE
His current research focuses on the development of novel optical imaging methods for investigating the role of the cellular microenvironment in disease, and the development of software for multidimensional image analysis. In 2008, he founded LOCI and started his research group at the University of Wisconsin-Madison.

Eliceiri completed his bachelor’s and doctoral degree in Biotechnology and Biomedical Engineering at the University of Wisconsin in Madison. Kevin Eliceiri is an Associate Professor of Medical Physics and Biomedical Engineering and director of the Laboratory for Optical and Computational Instrumentation (LOCI) at the University of Wisconsin-Madison. Second Speaker: Kevin Eliceiri (University of Wisconsin-Madison)ĭr. In 2017, Carpenter started her laboratory at the Broad Institute where she combines her background in cell biology, microscopy, and computational biology to develop methods extracting quantitative information from biological images. Polina Golland of MIT’s Computer Science/Artificial Intelligence Laboratory. David Sabatini and was co-mentored by Dr. She continued her scientific training as a postdoctoral fellow at the MIT/Whitehead Institute for Biomedical Research in the laboratory of Dr. Carpenter completed her bachelor’s degree in biological sciences, and a doctoral degree in cell biology from the University of Illinois, Urbana-Champaign.
THE INTENSITIES OF BANDS WERE DETERMINED BY IMAGEJ SOFTWARE SERIES
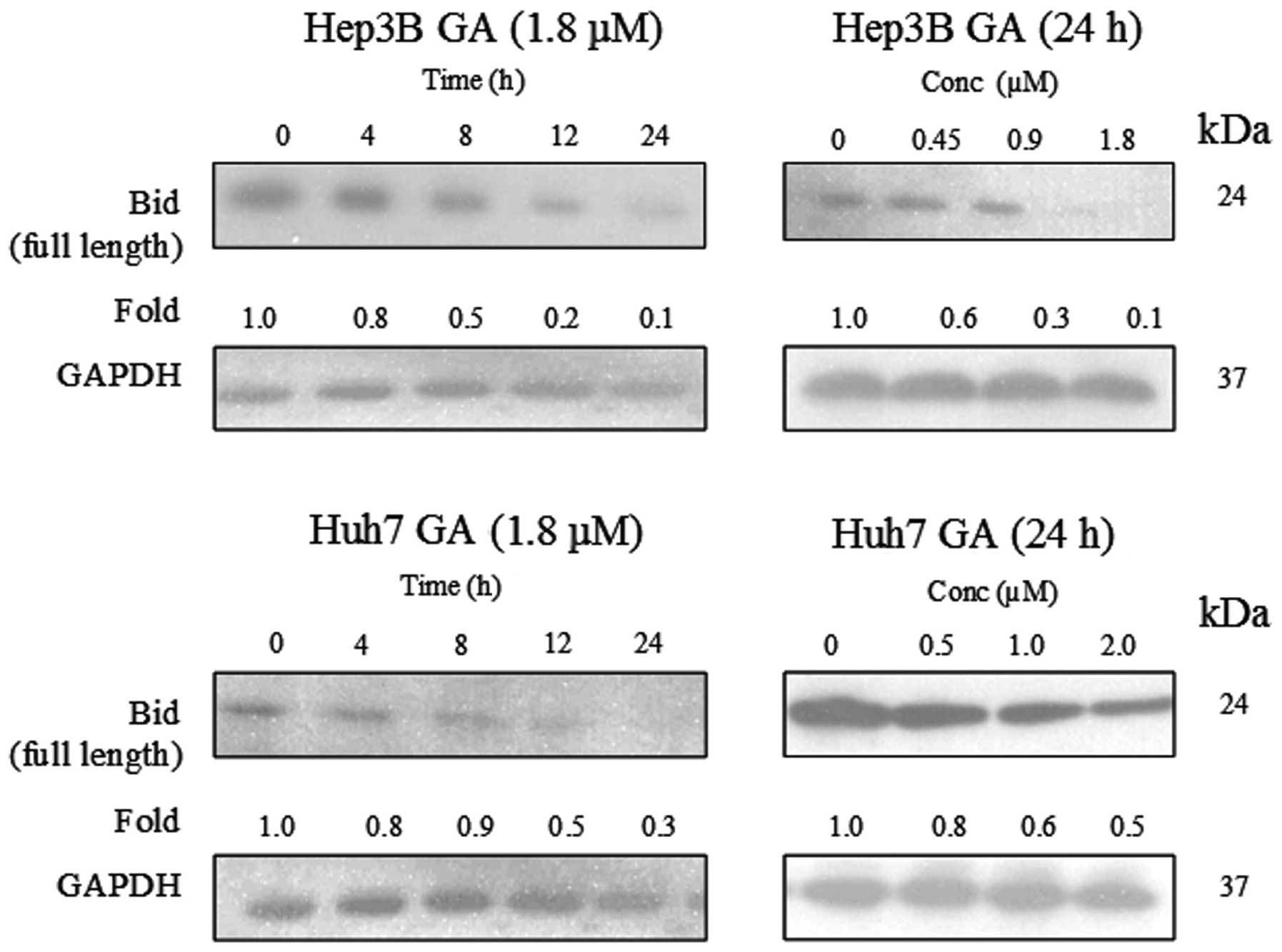
Carpenter and Eliceiri finalize this series by providing tips on best practices that will aid scientists in properly analyzing their data.įirst Speaker: Anne Carpenter (Broad Institute)Īnne Carpenter is an Institute Scientist at the Broad Institute of MIT and Harvard. The last step of bioimage analysis is to analyze the data by measuring different features like the number of cells or biological structures, or their size, shape, intensity or texture.

If appropriate for your dataset, you can use tracking to be able to link objects in space and time and measure speed, directionality, and cell division. Once you perform pre-processing, you’re ready for segmentation, the process of identifying individual cells or structures within an image.
Pre-processing is the first step that follows image acquisition and will prepare your image by reducing the signal-to-noise ratio, applying appropriate filters to the image, and color extraction. Kevin Eliceiri provide an overview of bioimage analysis.


 0 kommentar(er)
0 kommentar(er)
